We are quantifying sequence differences of entire genomes for individuals within and between populations and among species in order to understand the evolutionary processes underlying genome evolution. We are particularly interested in the interface between evolutionary forces — to characterize the interplay between natural selection, recombination, mutation, genetic drift, and population demography.
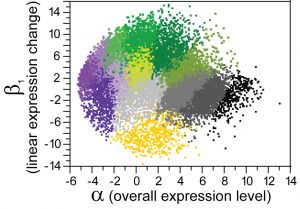 Our research aims to understand how adaptation and purifying selection alter the landscape of sequence variation and divergence across the genome (genetic hitchhiking, background selection), the extent to which crossover recombination and gene conversion affect natural selection, how different mutational processes impact inferences about selection and population history (demography, breeding systems), and the role of very weak natural selection on genomic features. We also use experiments measuring transcriptomes and allele-specific expression of coding sequences and small RNAs to help understand molecular evolution of gene regulatory controls. Example discoveries: C. brenneri has the highest molecular diversity of any animal or plant! Nematode miRNA genes and Argonaute gene families evolve in fascinating ways! Different ongoing forces and factors (recombination, mutation, selection, demography, breeding system, development) shape genetic variation and molecular evolution throughout genome! Our recent discoveries of new species and divergent populations makes genome-scale divergence population genetics analysis an exciting means of quantifying adaptation and other evolutionary processes with unprecedented resolution.
Our research aims to understand how adaptation and purifying selection alter the landscape of sequence variation and divergence across the genome (genetic hitchhiking, background selection), the extent to which crossover recombination and gene conversion affect natural selection, how different mutational processes impact inferences about selection and population history (demography, breeding systems), and the role of very weak natural selection on genomic features. We also use experiments measuring transcriptomes and allele-specific expression of coding sequences and small RNAs to help understand molecular evolution of gene regulatory controls. Example discoveries: C. brenneri has the highest molecular diversity of any animal or plant! Nematode miRNA genes and Argonaute gene families evolve in fascinating ways! Different ongoing forces and factors (recombination, mutation, selection, demography, breeding system, development) shape genetic variation and molecular evolution throughout genome! Our recent discoveries of new species and divergent populations makes genome-scale divergence population genetics analysis an exciting means of quantifying adaptation and other evolutionary processes with unprecedented resolution.