In the Cutter lab, we study the genetic basis of evolutionary change. We are particularly interested heritable changes through time with causes that are at the interface of natural selection and non-adaptive evolutionary forces. Our research currently centers on 3 broad themes: Population genomics and genome evolution, the genetics and developmental biology of speciation, and evolutionary cell biology of sperm.
We exploit C. elegans — the classic nematode roundworm model organism for genetics, development and neurobiology — and related species as a model to dissect these diverse problems in evolutionary biology. Caenorhabditis is an exceptional group to study the evolutionary process at molecular and phenotypic levels for many reasons: The genomes of Caenorhabditis species are modest in size and complexity (6 chromosomes, 50-150Mbp long, no genome duplications), species differ in effective population size (Ne: 104 – 107) and mode of reproduction (selfing hermaphrodites vs. dioecious separate sexes), behaviors and fitness may be quantified simply due to experimental tractability (1mm long, 3 day generation time, rearing on bacteria-spotted petri dishes), and abundant genetic and molecular tools from the C. elegans research community (RNAi, mutant/knockout alleles, CRISPR/Cas9 and other transgenic strains). We use these tools to help us link micro-evolutionary processes to macro-evolutionary pattern and molecular mechanisms.
Population Genetics and Genome Evolution
We are quantifying sequence differences of entire genomes for individuals within and between populations and among species in order to understand the evolutionary processes underlying genome evolution. We are particularly interested in the interface between evolutionary forces — to characterize the interplay between natural selection, recombination, mutation, genetic drift, and population demography.
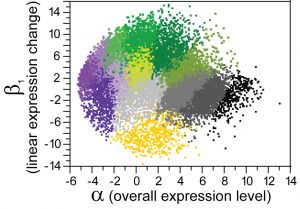 Our research aims to understand how adaptation and purifying selection alter the landscape of sequence variation and divergence across the genome (genetic hitchhiking, background selection), the extent to which crossover recombination and gene conversion affect natural selection, how different mutational processes impact inferences about selection and population history (demography, breeding systems), and the role of very weak natural selection on genomic features. We also use experiments measuring transcriptomes and allele-specific expression of coding sequences and small RNAs to help understand molecular evolution of gene regulatory controls. Example discoveries: C. brenneri has the highest molecular diversity of any animal or plant! Nematode miRNA genes and Argonaute gene families evolve in fascinating ways! Different ongoing forces and factors (recombination, mutation, selection, demography, breeding system, development) shape genetic variation and molecular evolution throughout genome! Our recent discoveries of new species and divergent populations makes genome-scale divergence population genetics analysis an exciting means of quantifying adaptation and other evolutionary processes with unprecedented resolution.
Our research aims to understand how adaptation and purifying selection alter the landscape of sequence variation and divergence across the genome (genetic hitchhiking, background selection), the extent to which crossover recombination and gene conversion affect natural selection, how different mutational processes impact inferences about selection and population history (demography, breeding systems), and the role of very weak natural selection on genomic features. We also use experiments measuring transcriptomes and allele-specific expression of coding sequences and small RNAs to help understand molecular evolution of gene regulatory controls. Example discoveries: C. brenneri has the highest molecular diversity of any animal or plant! Nematode miRNA genes and Argonaute gene families evolve in fascinating ways! Different ongoing forces and factors (recombination, mutation, selection, demography, breeding system, development) shape genetic variation and molecular evolution throughout genome! Our recent discoveries of new species and divergent populations makes genome-scale divergence population genetics analysis an exciting means of quantifying adaptation and other evolutionary processes with unprecedented resolution.
Genetics (and Development) of Speciation
We are capitalizing on the recent discovery of new Caenorhabditis species that are capable of forming inter-species hybrids to study the genetic basis of reproductive isolation — “speciation genetics.” To this end, we are addressing questions like: What governs the strong male inviability and sterility in inter-species hybrids (Haldane’s Rule)? What causes one species to be a better “mom” than another in inter-species hybrids (Darwin’s Corollary)? How much genetic variation within-species is there for between-species hybrid viability and fertility, and how fast does it accumulate over evolutionary time? How can gametes create reproductive isolation, and how can gamete incompatibilities influence the coexistence of species in community ecology? What genetic differences are responsible for reproductive isolation between species? What genetic and developmental mechanisms are responsible for inter-species incompatibility?
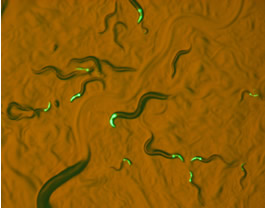 Our research brings the superb experimental power of Caenorhabditis to investigate the genetic basis of reproductive isolation in unprecedented ways. We are exploring this at several levels, from pre-mating factors like the evolution of male responses to mating pheromone to post-mating pre-zygotic factors implicating gametic isolating barriers to post-zygotic incompatibilities that cause inviability and sterility in hybrids. Using powerful tools, including inter-species NIL mapping, in vivo sperm labeling, and genome editing, we are dissecting the mechanistic and genetic causes of these barriers to gene flow. We are especially interested in integrating the genetics of speciation with the development over ontogeny to connect evo-devo and speciation genetics.
Our research brings the superb experimental power of Caenorhabditis to investigate the genetic basis of reproductive isolation in unprecedented ways. We are exploring this at several levels, from pre-mating factors like the evolution of male responses to mating pheromone to post-mating pre-zygotic factors implicating gametic isolating barriers to post-zygotic incompatibilities that cause inviability and sterility in hybrids. Using powerful tools, including inter-species NIL mapping, in vivo sperm labeling, and genome editing, we are dissecting the mechanistic and genetic causes of these barriers to gene flow. We are especially interested in integrating the genetics of speciation with the development over ontogeny to connect evo-devo and speciation genetics.
Evolutionary Cell Biology of Sperm
Evolution of gametic cells provides a microcosm of the interface between natural selection and sexual selection, including sexual conflict between the fitness interests of the sexes. We found Caenorhabditis‘ amoeboid sperm cells to have evolved radical size diversification, with a >50-fold range in volume across species. What other fitness-related traits associate with sperm size gigantism across phylogeny? What implications does sperm gigantism have for sexual selection and sexual conflict? Does harmful gamete-mediated reproductive interference affect multi-species community composition? How do subcellular sperm traits relate to cell size? How does temperature influence sperm cell fecundity?
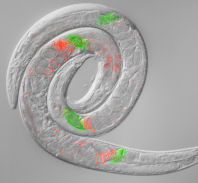 Shifts in reproductive mode have driven drastic changes in the genes responsible for sperm competition. The sperm gigantism of Caenorhabditis has led us to use electron microscopy to peer inside these cells with even greater resolution, and the ectopic invasiveness of sperm cells suggests that they may provide a mechanism of reproductive interference competition between species. We are documenting temperature-dependent traits that differ between latitudinally– and phylogeographically-distinct strains of C. briggsae. We have interrogated the genetic basis of temperature-related traits with recombinant inbred lines (RILs) to QTL map natural phenotypic differentiation thought to reflect local adaptation. We also are quantifying transcriptome responses to temperature and genotype.
Shifts in reproductive mode have driven drastic changes in the genes responsible for sperm competition. The sperm gigantism of Caenorhabditis has led us to use electron microscopy to peer inside these cells with even greater resolution, and the ectopic invasiveness of sperm cells suggests that they may provide a mechanism of reproductive interference competition between species. We are documenting temperature-dependent traits that differ between latitudinally– and phylogeographically-distinct strains of C. briggsae. We have interrogated the genetic basis of temperature-related traits with recombinant inbred lines (RILs) to QTL map natural phenotypic differentiation thought to reflect local adaptation. We also are quantifying transcriptome responses to temperature and genotype.